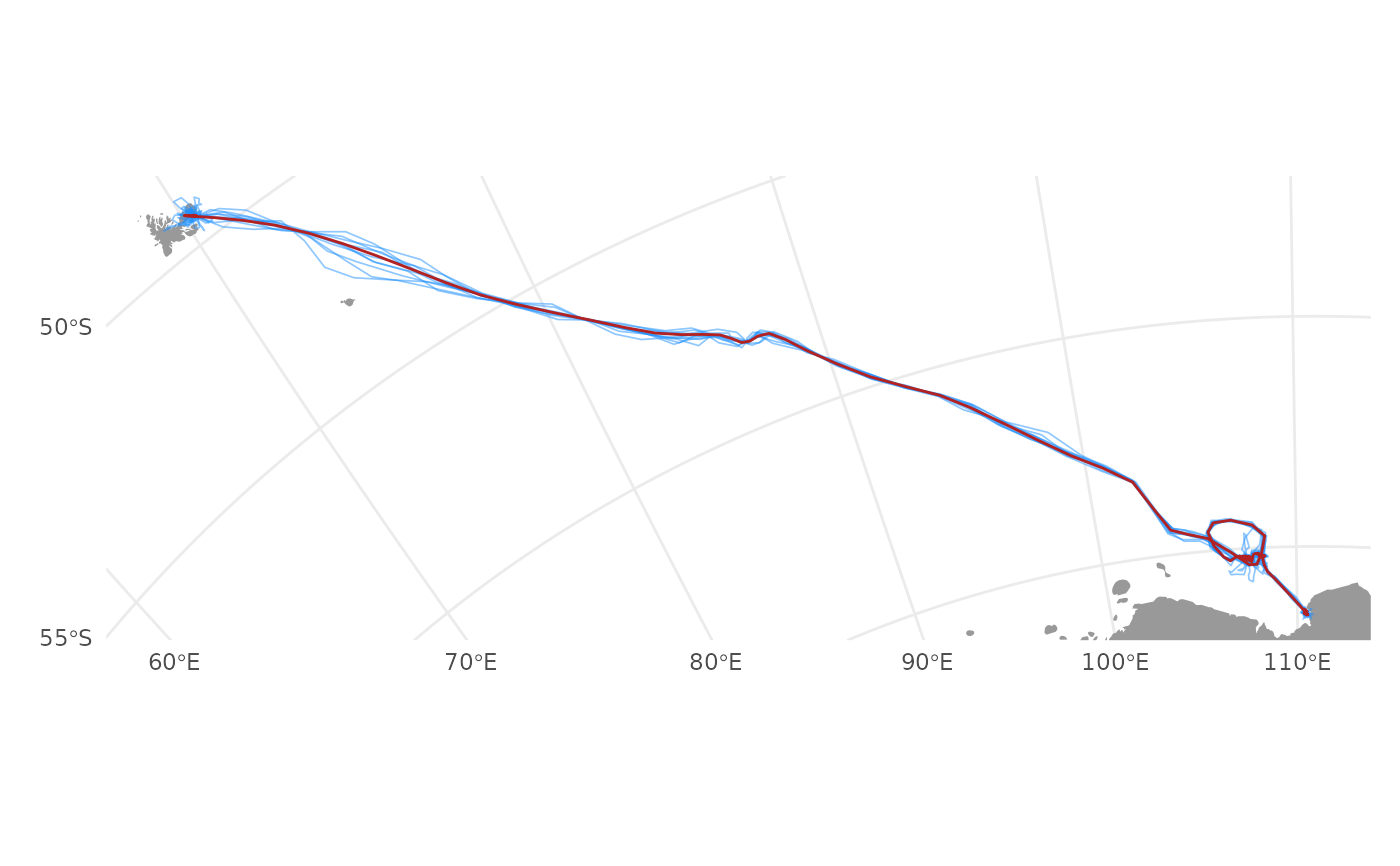
visualize tracks simulated from a aniMotum model fit
Arguments
- x
a
aniMotumsimulation data.frame with classsim_fit- type
plots tracks as "line", "points" or "both" (default).
- zoom
logical; should map extent be defined by track extent (TRUE; default) or should global map be drawn (FALSE).
- ncol
number of columns to arrange multiple plots
- hires
logical; use high-resolution coastline data. Attempts to use high-res coastline data via rnaturalearth::ne_countries with
scale = 10, if thernaturalearthhiresdata package is installed. If not, then rnaturalearth::ne_countries withscale = 50data are used.- ortho
logical; use an orthographic projection centered on the track starting location(s) (TRUE; default). An orthographic projection may be optimal for high latitude tracks and/or tracks that traverse long distances. If FALSE then a global Mercator projection is used.
- alpha
opacity of simulated track points/lines. Lower opacity can ease visualization when multiple simulated overlap one another.
- ...
additional arguments to be ignored
Value
Plots of posterior simulated tracks.
Examples
fit <- fit_ssm(ellie, model = "crw", time.step = 24)
#> fitting crw SSM to 1 tracks...
#>
pars: 1 1 0 -2.91153
pars: 0.38512 0.37191 -0.01692 -3.38813
pars: -1.4595 -1.51236 -0.06769 -4.81792
pars: -2.20058 -2.24383 -0.11527 -4.95732
pars: -0.38346 -0.4056 -0.04615 -3.85456
pars: -1.23664 -1.20844 -0.10327 -3.65984
pars: -2.25711 -2.16871 -0.17159 -3.42693
pars: -3.07868 -3.34599 -0.26131 -1.24778
pars: -4.46619 -1.62751 -0.49092 0.12579
pars: -3.18442 -2.86245 -0.30197 -0.80151
pars: -3.40955 -3.30581 -0.49919 -0.40195
pars: -3.06893 -3.16418 -1.05454 -0.36585
pars: -3.5513 -2.81435 -1.34642 -0.44033
pars: -3.30092 -3.01944 -1.10395 -0.39263
pars: -3.27833 -3.03821 -1.38152 -0.39631
pars: -3.23173 -2.97174 -2.21731 -0.40477
pars: -3.28771 -3.08271 -5.28966 -0.34232
pars: -3.30222 -3.07091 -5.31779 -0.38305
pars: -3.30222 -3.07091 -5.31779 -0.38305
psim <- sim_post(fit, what = "p", reps = 10)
plot(psim, type = "lines")
#> Warning: `label` cannot be a <ggplot2::element_blank> object.
#> Warning: `label` cannot be a <ggplot2::element_blank> object.