This function calculates the similarity between the simulations
generated by sim_fit and the SSM-estimated path from the ssm fit,
and returns a sim_fit object containing the most similar tracks based on
a user specified quantile. In this context, similarity is calculated
as the sum of normalised differences in net displacement (km) and overall
bearing (deg) between the SSM-estimated path and the simulated paths.
sim_filter(trs, keep = 0.25, flag = 2, var = NULL, FUN = "mean", ...)Arguments
- trs
a
sim_fitobject- keep
the quantile of flag values to retain
- flag
the similarity flag method (see details). Ignored if var != NULL.
- var
the name(s) of the appended variable(s) to use for similarity calculations. Default is NULL, in which case similarity is calculated based on distance and bearing - e.g., Hazen et al (2017).
- FUN
one of the following functions in quotes: mean, median, var, sd, sum, min, or max. Ignored if var = NULL.
- ...
additional arguments to the specified FUN (e.g., na.rm = TRUE). Ignored if var = NULL.
Value
a sim_fit object containing the filtered paths
Details
flag = 1will use an index based on Hazen (2017)flag = 2(the default) will use a custom index
References
Hazen et al. (2017) WhaleWatch: a dynamic management tool for predicting blue whale density in the California Current J. Appl. Ecol. 54: 1415-1428
Examples
## fit crw model to Argos LS data
fit <- fit_ssm(ellie, model = "crw", time.step = 72)
#> fitting crw SSM to 1 tracks...
#>
pars: 1 1 0 -3.35398
pars: 0.39011 0.36995 -0.01715 -3.83437
pars: -1.43958 -1.5202 -0.06862 -5.27553
pars: -2.32555 -2.54729 -0.19371 -4.12372
pars: -0.8182 -0.89535 -0.06643 -4.50381
pars: -1.32518 -1.50336 -0.13293 -2.80663
pars: -2.70765 -2.68894 -0.23564 -2.37746
pars: -3.27563 -3.39761 -0.40156 -0.74674
pars: -4.54561 -2.25375 -0.86806 -0.13627
pars: -3.3297 -3.27793 -0.43777 -0.61077
pars: -3.36938 -3.29518 -0.51676 -0.44068
pars: -3.37129 -3.28379 -0.68713 -0.35188
pars: -3.37129 -3.28379 -0.68713 -0.35188
set.seed(pi)
## generate 5 simulated paths from ssm fit
trs <- sim_fit(fit, what = "predicted", reps = 5)
## filter simulations and keep paths in top 40% of flag values
trs_f <- sim_filter(trs, keep = 0.4, flag = 2)
## compare unfiltered and filtered simulated paths
# \donttest{
plot(trs) | plot(trs_f)
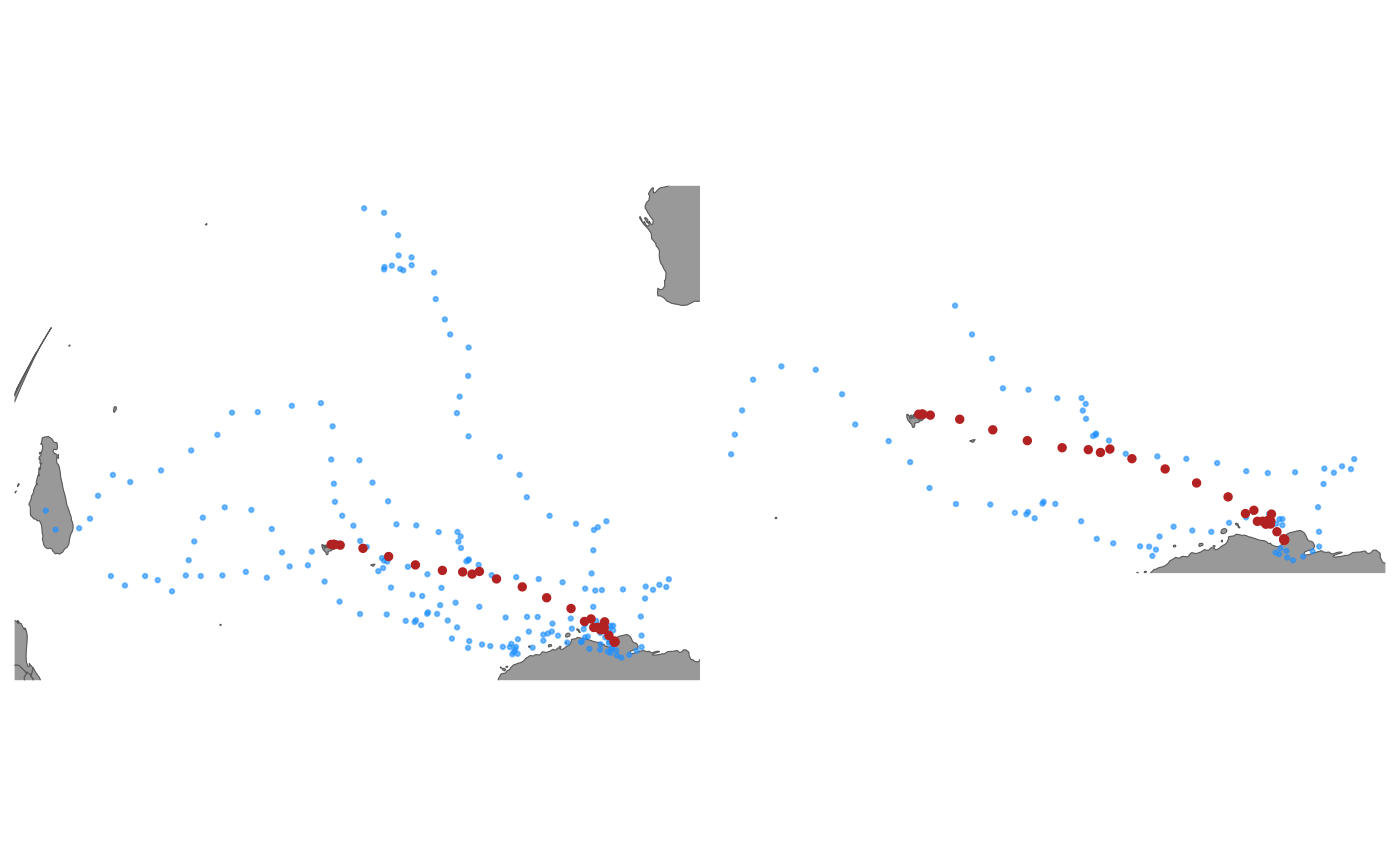 # }
# }